Research
Research Projects & Grants
Histopathology: Diagnosing diseases from histopathology slides can be subjective and time-consuming. We are developing explainable AI methods highlighting regions of interest for pathologists, helping speed up diagnosis and reduce subjectivity in interpretation.
Radiology: Our research spans various topics, including medical image registration and tumor segmentation in ultrasound and CT images. Collecting and annotating large datasets for training deep learning models is often costly and requires expert radiologists. To address this, we are developing weakly supervised methods for tumor segmentation in CT scans. We are also exploring the use of synthetic data, generated using GANs and diffusion models to improve segmentation performance. This is especially beneficial in smaller countries, where collecting large-scale medical datasets is challenging due to limited population size.
Microscopy: We focus on instance segmentation and classification of tumors using brightfield microscopy images. There is a lack of publicly available datasets for instance segmentation, particularly for overlapping cases. To address this, we have curated and released a dataset that includes such challenging cases. Additionally, we are developing advanced end-to-end architectures for instance segmentation. Other areas of our work include identifying and segmenting cytopathic effects and classifying cell cycle stages using microscopy images.
Below are some of the recent projects from our lab.
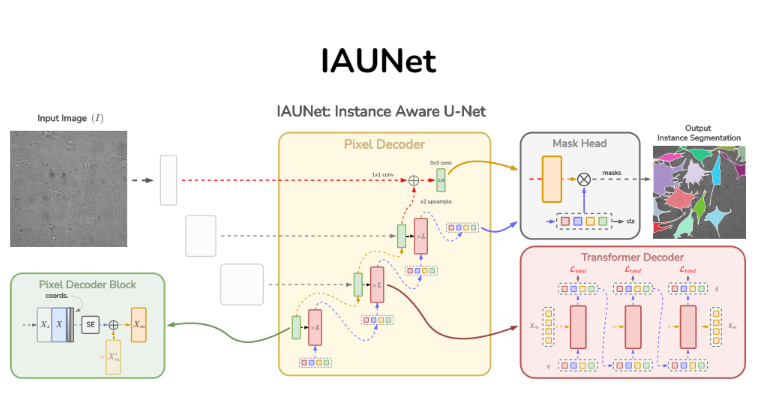
Instance-aware segmentation of microscopy images
Instance segmentation is vital in biomedical imaging for accurately separating overlapping objects like cells. We propose IAUNet, a novel query-based U-Net architecture that retains U-Net’s structure while introducing a lightweight Pixel decoder and a Transformer decoder with deep supervision. IAUNet efficiently combines multi-scale features and refines object-specific queries for precise instance segmentation. Experiments on multiple datasets demonstrate that IAUNet outperforms state-of-the-art convolutional, transformer-based, and cell-specific segmentation models.
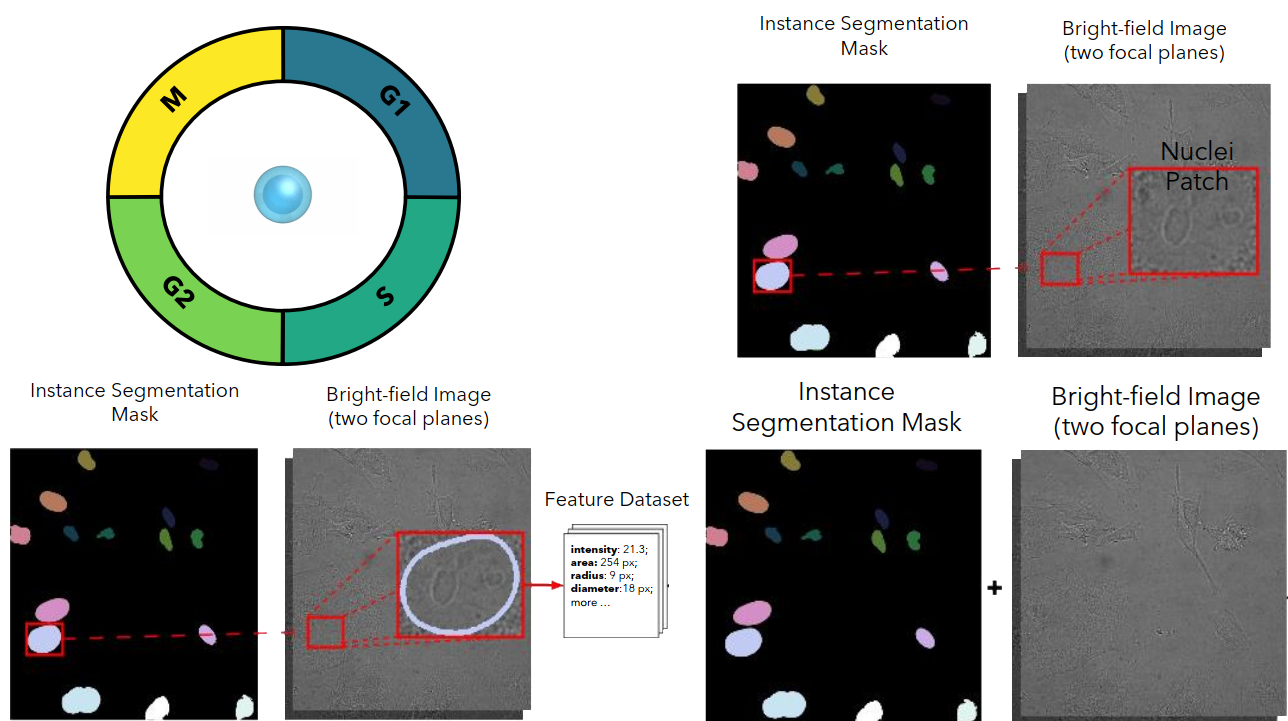
Cell Cycle Phase Classification from Microscopy Images
Accurate cell cycle phase classification is vital for cancer research and drug discovery. This study evaluates five computational strategies using fluorescence and bright-field microscopy. While fluorescence enables near-perfect performance, the results demonstrate that deep learning methods can also achieve strong accuracy with label-free bright-field images, paving the way for scalable biomedical applications.
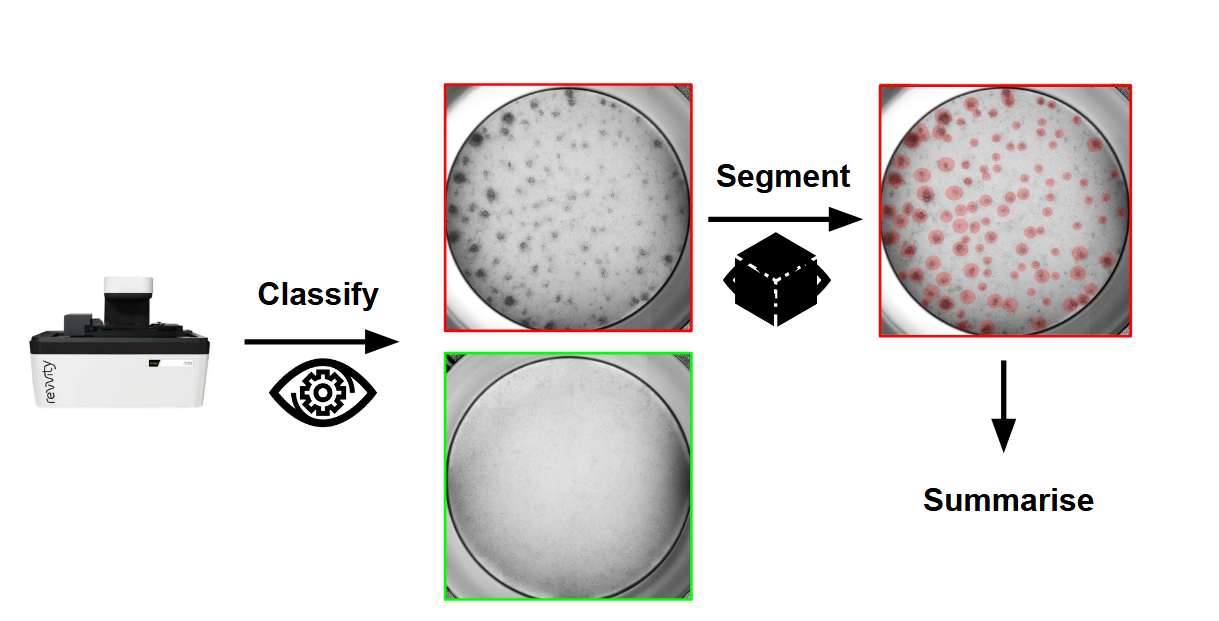
Mastering the Unseen: Approaches to Hard-to-Detect Viral Cytopathic Effect
Cytopathic Effect (CPE) serves as a key visual indicator of viral infection but is challenging to assess manually. This study evaluates classical and deep learning-based computer vision methods, ranging from classification to weakly and strongly supervised segmentation, for automated CPE detection from x-MuLV-infected cells. Results show that supervised segmentation provides more reliable quantification, especially in subtle or low viral load cases, enabling scalable, objective, and high-throughput viral analysis. This work identifies best-performing approaches to support drug discovery and virological research.
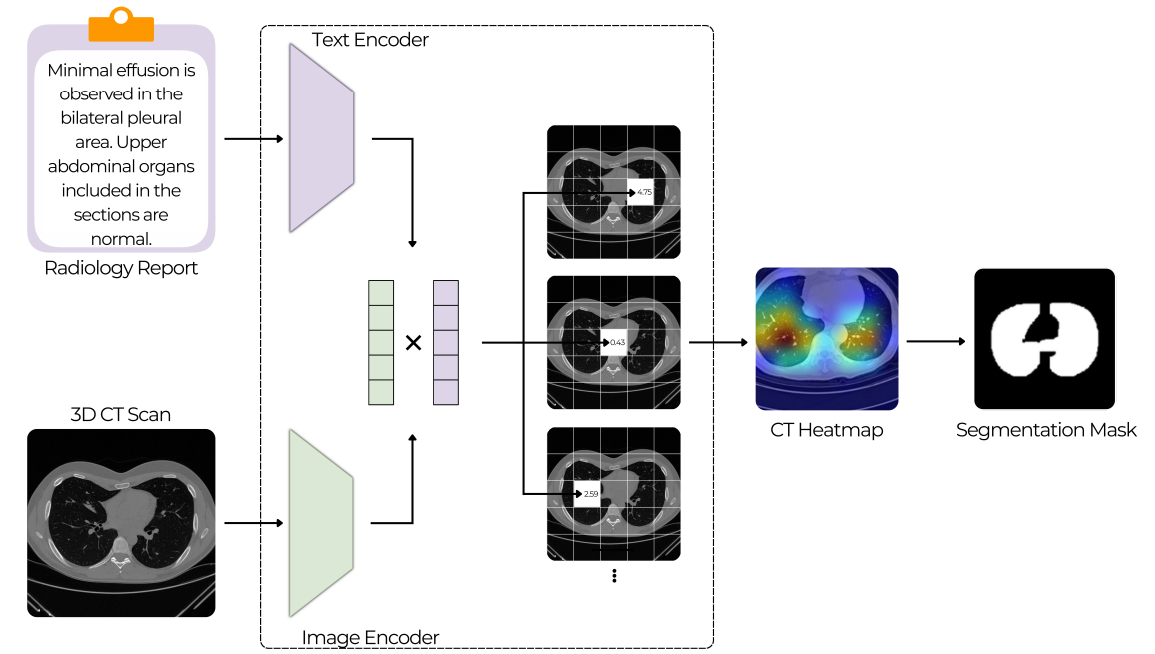
Text-Driven Weakly Supervised Medical Image Segmentation
This work explores the use of multimodal vision-language models for medical image analysis, focusing on 3D CT scans paired with radiology reports. Unlike traditional CNNs, these models can leverage textual context to potentially learn spatial localization without explicit segmentation labels. The study investigates whether such models exhibit emergent segmentation abilities, offering a scalable solution to the limited availability of annotated medical data and supporting clinical workflows through weak supervision.
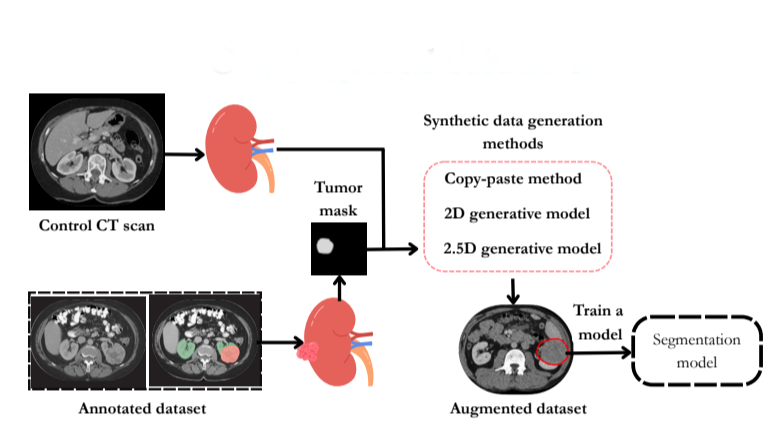
Evaluating the Impact of Synthetic Data on Enhancing Kidney Tumor Segmentation
This study addresses the challenge of kidney tumor segmentation by exploring three synthetic data generation methods: copy-paste augmentation, 2D diffusion, and 2.5D diffusion models. Results show that synthetic data, particularly from copy-paste augmentation, improves segmentation performance despite limited annotations. Although diffusion models offer better context, they need refinement to be competitive. This work highlights the potential of synthetic data to enhance clinical AI segmentation systems.
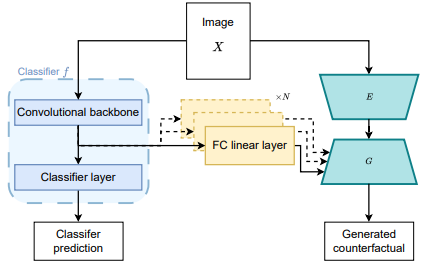
Improving Counterfactual Image Generation for Weakly Supervised Tumour Segmentation through Theoretical and Architectural Alignment
This work investigates architectural improvements in weakly supervised tumor segmentation using a classifier-guided conditional GAN framework with image-level labels. Key modifications, including removing bias, integrating classifier features, and aligning discriminator training, were tested on the TUH kidney tumor CT dataset. Results show comparable segmentation accuracy to baselines, with reduced constraints and training time cut by up to two-thirds, highlighting the benefits of architectural alignment in weak supervision.
